Research Article 
 Creative Commons, CC-BY
Creative Commons, CC-BY
Random Forest-Based Compound ATC Classification Using Structural and Physiochemical Information
*Corresponding author: Marcel Angelo Daniel Gunadi, De Anza college, USA.
Received:October 18, 2022; Published: October 26, 2022
DOI: 10.34297/AJBSR.2022.7.002344
Abstract
There are nearly 4,000 FDA-approved drugs (including salts) for different indications. An additional 4,600 compounds have an ongoing investigation in clinical trials. These drugs and compounds represent a mere fraction of the total chemical space, approximated to be greater than 10e12. Approximately 1.8 million compounds from the chemical space represent potential candidates for FDA approval and are currently being classified via traditional experimental methods, which are both costly and laborious, yet notalways- deterministic. This highlights the need for automated Anatomical Therapeutic Chemical (ATC) classification, which provides classification systems for drugs or drug-like substances into five levels. ATC level one and level two classifications are available only for 2,739 compounds (in ChEMBL). In this research, two random forest-based ensemble models are trained to predict the ATC classes for the 1.8M preclinical compounds available at ChEMBL. We used structural and physiochemical properties for the feature extraction. Using independent testing, we obtained a micro F1 score of 0.69 and 0.81 for the ATC level one and two models respectively, with the ATC level one model showing a greater accuracy (71.9%) compared to existing ATC level one classification methods. This research illustrates how improved classification of preclinical compounds may enhance the performance of future in-silico-based drug repurposing methods and help understand the mode of action for the preclinical compounds.
Keywords: Drug repurposing, Compound classification, ATC classification, Anatomical Therapeutic Chemical classification, Drug mode of action, Random forest
Introduction
The World Health Organization Collaborating Center (WHOCC) developed the Anatomical Therapeutic Chemical (ATC) system to classify drugs or drug-like substances. It is one of the most widely used systems for classifying compounds, dividing them into several classes at five levels [1]. Each level of ATC classification represents a specific mode of classification. ATC level one groups compounds into 14 classes based on anatomical properties, each represented by an English capital letter. ATC level two groups’ compounds into 94 classes based on therapeutic effects, each represented by two digits. Level three groups’ compounds pharmacologically into 267 classes and is represented by an English letter. Level four and five are based on chemical properties and have 889 and 5,056 classes, respectively. More details on ATC classification systems are available at: https:// www.whocc.no/atc_ddd_index/. Presently, only a few thousand compounds have ATC classifications at a certain level. WHOCC is responsible for assigning ATC classification if drug manufacturing organizations or pharmaceutical companies have requested it. However, the current process is cumbersome, failing to leverage the capabilities of modern technology to maximize efficiency. With the advent of high throughput technologies, millions of compounds are now publicly available [2,3]. Hence, there is a need to develop machine learning (ML) based prediction models to assign classes for newly screened compounds automatically.
Several ML-based models have been proposed to predict ATC classes for the compounds in the recent decade. Dunkel, et al. [1] developed a logistic regression-based model for ATC prediction using chemical structural features. They also provided a publicly available web server (SuperPred) to predict disease indications based on the properties and similarities of compounds. The iSEA (indication similarity ensemble approach), a logistic regression model, utilized chemical information, target proteins, gene expressions, and side effect profiles to predict ATC codes [4]. Chen, et al. [5] used chemical-chemical interactions to predict ATC classes using statistical approaches. Another study [6] was based on natural language processing techniques to predict ATC class labels. The iATC-mHyb [7] is a multi-label Gaussian kernel regression (ML-GKR) based predictor that classifies compounds into ATC level one classes. The iATC-mISF [8] also used ML-GKR trained on compound-compound interactions and structural similarities to predict the ATC level one class of the compounds.
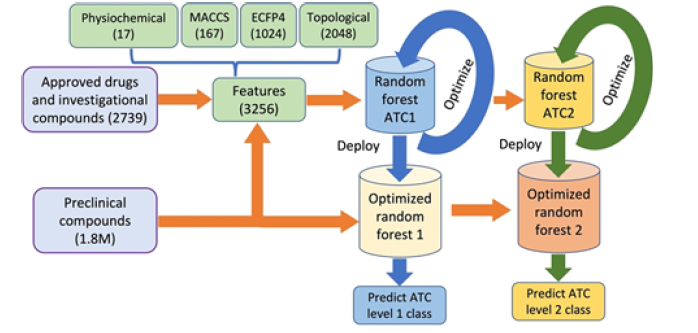
The compound classification models, as reported above, are appropriate for the ATC classification of compounds; however, to our knowledge, none have applied their models to the classification of millions of preclinical compounds available in PubChem [9] and ChEMBL [10] databases. Furthermore, the aforementioned models are not based on comprehensive structural and physiochemical features as adapted in the proposed research. In this research, we proposed two random forest-based ensemble models to be trained on ATC-classified compounds available at ChEMBL. The first random forest classifier is trained to predict ATC level one classes and the second to predict ATC level two classes. The proposed random forest classifiers are based on structural and physiochemical features. To represent the structural features of compounds, we combined three molecular fingerprints: 1) Molecular ACCess System (MACCS) [11], 2) Extended-connectivity fingerprints (ECFP4) [12], and 3) topological fingerprints [13]. After successful training, we applied the two trained random forest classifiers to predict classes for ATC levels one and two for 1.8M preclinical compounds. The prediction of ATC classes for preclinical compounds may help to understand possible side effects, improve structure–function prediction, and speed up the drug discovery process.
Materials and Methods
Gold Standard Dataset
ChEMBL is one the most comprehensive datasets providing ATC classifications for compounds up to five levels. By querying the ChEMBL database, we extracted 2,739 approved and investigational compounds for which ATC level one and level two classifications were available. Of these 2,739 compounds, 2,516 were approved small molecules (or salts), 169 were in phase 3, 44 were in phases 2, and 10 were in phase 1, as shown in Figure 1. As mentioned in the introduction, ATC level two contains 94 classes. However, the ChEMBL database contains compounds for only 87 out of the 94 classes. Therefore, we could train the proposed ATC level 2 model only for the 87 classes.
We used one-hot encodings to map ATC class names from text into numeric. The ATC level one classes have numeric labels 1-14. Similarly, the ATC level two classes have numeric labels 1-87. The prediction algorithm is trained and optimized on this gold standard dataset and later applied to the 1.8M Preclinical compounds. Each compound is uniquely represented by structural identifiers such as the Standard InChiKeys and Smiles, which are also extracted from ChEMBL (Figure 1).
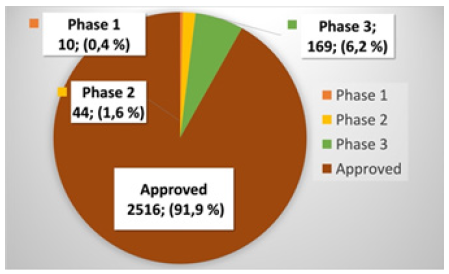
Figure 1: Distribution of drugs and investigational compounds by the maximum clinical phases against any indication.
Feature Extraction
The ML algorithms are trained on numeric data; therefore, we computed the numerical features for the 2,739 training and 1.8M preclinical compounds. Two types of features are computed, i.e., physiochemical and structural features (using fingerprints) for the compounds. Details of these feature extraction methods are given in the following sections.
Physiochemical features: We computed 17 such
physiochemical features listed below:
i. Molecular weight of parent compound
ii. AlogP
iii. Number of hydrogen bond acceptors
iv. Number of hydrogen bond donors
v. Polar surface area
vi. Number of rotatable bonds
vii. Number of violations of Lipinski’s rule-of-five
viii. Most acidic pKa calculated using ChemAxon
ix. Calculated octanol/water partition coefficient using
ChemAxon
x. Calculated octanol/water distribution coefficient at pH7.4
xi. Molecular weight of the full compound
xii. Number of aromatic rings
xiii. Number of heavy atoms
xiv. Weighted quantitative estimate of drug-likeness
xv. Monoisotopic parent molecular weight
xvi. Number of hydrogen bond acceptors calculated according
to Lipinski’s original rules
xvii. Number of hydrogen bond donors calculated according to
Lipinski’s original rules.
These 17 physiochemical features are computed using the RDKit [14] python package by inputting the smiles of a compound. The physiochemical properties are computed for each of 2,739 training and 1.8M preclinical compounds.
Structural features: We computed structural features by
combining three molecular fingerprints, i.e., Molecular ACCess
System (MACCS), Extended-connectivity fingerprints (ECFP4), and
topological fingerprints, as mentioned in the introduction. These
three fingerprints are also computed using RDKit by inputting the
smiles of the compounds. The structural properties are computed
for 2,739 training and 1.8M preclinical compounds. The smiles of
the 2,739 training compounds and 1.8M preclinical compounds
are downloaded from the ChEMBL database, as mentioned in
section 2.1. The fingerprint is a vector of binary numbers that
represent the sub-structures present in the compounds (1 for
the presence and 0 for the absence of a particular sub-structure).
The MACCS, ECFP4, and topological fingerprint lengths are 167,
1,024, and 2,048, respectively. Therefore, the combined structural
feature representation for each compound is of length: 3,239
(167+1024+2048=3239).
We combined the 3,239 structural features with 17
physiochemical properties. Therefore, the combined size of the
feature vector is 3,256 features for each compound.
Training of Prediction Models
ATC levels one and two have several classes; we, therefore, formulated this problem as a multi-classification problem, for which a random forest was applied. The random forest, developed in 1995 [15], is an ensemble classification technique based on multiple decision trees. Each decision tree predicts a class label and then assigns them. The random forest then assigns the class labels based on the majority voting of predictions by the individual decision trees. The strength of each tree and the correlation in between determine the generalization error of the random forest [16]. Due to the combined prediction power of individual decision trees, random forests tend to have better prediction accuracies than other classification algorithms [17]. Secondly, random forests are frequently used in drug discovery [18] and development studies and are highly successful for different tasks such as drug sensitivity prediction [19] and drug target prediction [19,20]. We, therefore, adapted a random forest-based ensemble model in this research. As we are predicting classes for ATC level one and level two, we trained two random forest-based ensemble models based on 3,256 features. We also computed the same 3,256 features for 1.8M preclinical compounds. Like any other ML algorithm, random forest models involve hyperparameters to be optimized to achieve improved model performance [21].
Therefore, we tuned and optimized the hyperparameters of both random forest models. After successful training and optimization, the two random forest models were deployed to predict the ATC level one and level two classes of the unclassified compounds. Figure 2 shows the flow chart of the proposed methodology (Figure 2).
The 3,256 features are computed for the training and 1.8M preclinical testing compounds. The two random forest models (Random Forest ATC1 and Random Forest ATC2) are trained and optimized on the 2,739 approved drugs (and investigational compounds). After successful training, the two models are deployed to predict ATC level one and two classes of the 1.8M preclinical compounds.
Distribution of Compounds Across ATC Level One and Level Two Classes
The ChEMBL database contains ATC level one class labels for all 2,739 training compounds. There are 14 classes in ATC level one and 94 in ATC level two. However, ATC level two class labels are available only for 87 (out of 94) classes, as explained in section 2.1. For ATC level one, we have >40 compounds for each class, but the distribution of compounds/drugs across 14 level one classes is not uniform (Figure 3A). For uniform distribution, the average number of compounds per class should be 195.6 (2,739/14=195.6). In our dataset, six classes have less than 195 compounds. The training data is skewed, as seen in Figure 3A. Similarly, the data for ATC level two is even more skewed, as shown in Figure 3B. For uniform distribution, there should be 31.4 (2739/87=31.4) compounds for each class; however, more than 60 classes contain less than 31 compounds (Figure 3).
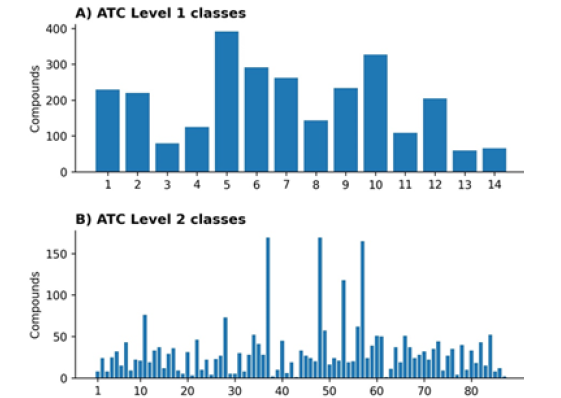
Figure 3: Distribution of compounds across different classes. A) ATC level one classes, B) ATC level two classes.
As the training datasets for ATC levels one and two are highly skewed, prediction algorithms can be biased towards the bigger classes. Possible solutions to counter this are to use either undersampling or over-sampling [22]. In this study, we adapted the synthetic minority over-sampling technique (SMOTE) [23] to oversample level one and two classes in the minority. SMOTE is a successful over-sampling technique for drug discovery applications [24,25]. After SMOTE based oversampling, we obtained 5,449 ATC level one compounds and 11,142 ATC level two compounds. These oversampled datasets are used to train and assess the performance of the proposed random forest classifiers for each level.
We used micro F1, micro precision, and micro recall metrics to assess the performance of the proposed random forest classifiers. The micro F1, micro recall, and micro precision scores are defined using the following equations.
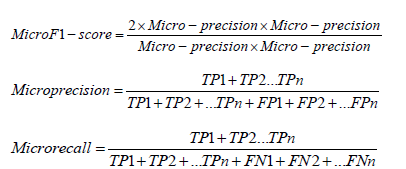
Results and Discussion
Hyperparameter Optimization
Every prediction algorithm has some hyperparameters
that need to be optimized for the given dataset to improve the
algorithm’s performance. These parameters can be optimized in
various ways. We adopted a random search algorithm [26] in this
research to find the optimal hyperparameters for both random
forest classifiers. The optimal values for the five hyperparameters
are shown as follows:
a. Number of decision trees in random forests = 200
b. Maximum features number = square root of the total
number of features
c. Maximum depth = 16
d. Minimum sample leaf = 2
e. Minimum sample split = 2
After computing the variables’ optimal settings, we tested the experiments, as explained in the following section.
Results for Testing Prediction Algorithms
As mentioned in the methods section, we trained two random forest classifiers, one for each level of the ATC classification system. We assessed the performance of the two classifiers using independent testing. Because of the SMOTE-based oversampling technique, we were able to avoid bias towards any individual class. A direct consequence of this technique is that there were duplicate compounds in smaller classes. After SMOTE based oversampling, the new size of the datasets was 5,449 and 11,142 compounds for ATC level one and level two, respectively.
To test the accuracy of our two random forest models, we divided both datasets of classified compounds into training and testing sets. For each dataset, we randomly picked 90% of the compounds for the training of classifiers and left 10% for testing to assess the performance of the two models. As a multi-classification problem, we assessed the performances of the classifiers based on micro F1, micro recall, and micro precision as defined in section 2.4..
Using independent testing for the ATC level one classification, we obtained micro F1 of 0.69, micro recall of 0.70, and micro precision of 0.68. For the ATC level two classifier, we obtained a micro F1, micro recall, and micro precisions of 0.81, 0.8, and 0.81, respectively, as shown in Table 1. The micro F1 for ATC level two classification is better than level one; one reason could be that compounds under ATC level two are easy to distinguish compared to level one classification. However, both models show reasonably better statistics as micro F1 of > 0.65 is generally considered an appropriate statistic for multi-class prediction algorithms [27]. Therefore, we can claim that the proposed random forest-based models can successfully predict ATC level and level two classes. After testing the accuracy of our model, we deployed both classifiers to predict ATC level one and two classes for the unknown compounds (Table 1).
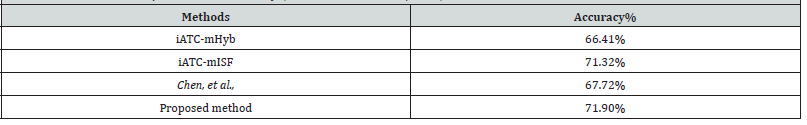
Comparison with Other Methods
We compared the proposed ATC level one model with the iATC-mHyb [7], iATC-mISF [8], and Chen, et al., [5]. We compared the results only for ATC level one due to the lack of methods for predicting ATC level two classes. Furthermore, we chose these methods for comparison because all three methods are tested on the same datasets, making it possible for us to perform direct comparisons. The three methods are trained on 3,883 compounds from the ChEMBL drug ontology database. Both iATC-mHyb and iATC-mISF are based on multi-label gaussian kernel regression. The method proposed by Chen, et al. is based on compound-compound interactions and structural information. All three methods used the accuracy metric to assess the performance. We applied the proposed level one predictor to predict the ATC level one classes for the same set of compounds used in the three methods. As shown in Table 2, our proposed random forest model outperformed the other three methods by achieving an accuracy of 71.9%. The better performance is due to the adoption of comprehensive structural and physiochemical properties. Each of the three structural fingerprints brought additional discriminant capabilities to the random forest models enabling a more comprehensive representation of the training data set, thus allowing the models to learn more effectively. Furthermore, these results reinforce the notion that a random forest achieves greater accuracy than the aforementioned models due to the combined prediction power of individual decision trees. Random forests, in general, are powerful ensemble models that can outperform traditional machine learning methods (Table 2).
Predicting Classes for ATC Level One and Two for 1.8M Preclinical Compounds
After successful training, we deployed the two models to predict ATC level one and level two classes for the 1.8M preclinical compounds. We provided the levels one and two predictions in Supplementary file 1. Supplementary file 1 contains three columns, 1) the Standard in Chi Key of the preclinical compound, 2) ATC Level 1 class, and 3) ATC level two class. The ATC classes for those 1.8M preclinical compounds are not available in any public resource. Therefore, it is not possible to validate our predictions. However, our predictions are valuable addition to drug discovery research and may help future in-silico-based models obtain higher prediction performance.
Conclusion
In this research, we proposed random forest-based classification algorithms to predict the ATC level one and two classes of compounds. We used comprehensive features derived from several structural and physiochemical properties. This comprehensive collection of features helped the random forestbased ensemble algorithms learn and successfully predict the ATC classes with a micro F1 of 0.69 and 0.81 for levels one and two, respectively. Compared with the iATC-mHyb, iATC-mISF and Chen, et al. methods, the proposed model performed slightly better, showing its effectiveness on unseen data. This could be due to the implementation of the effective feature sets to represent a particular compound. Finally, we applied the trained classifiers to predict ATC classes for 1.8M preclinical compounds, which is provided in Supplementary file 1. One limitation of the proposed method is that our ATC level 2 predictor can only classify compounds into 87 (out of the 94 classes). None of the 1.8M preclinical compounds or other compounds are classified into the remaining 7 ATC level two classes. This is because the classification algorithm for ATC level two was trained only on 87 classes (due to a lack of data).
Nonetheless, we believe that the predicted ATC classes for 1.8M preclinical compounds will help to understand the mode of action for these compounds and provide knowledge on possible adverse effects. Furthermore, it can help boost the performance of future compound-compound interaction and compound-protein interaction-based ML methods resulting in new drug repurposing applications. In future works, researchers may try to validate the predicted ATC classes for 1.8M preclinical compounds.
Conflict of Interest
Author doesn’t have any conflict of interests.
References
- M Dunkel, S Günther, J Ahmed, B Wittig, R Preissner (2008) SuperPred: drug classification and target prediction. Nucleic Acids Res 36(2): W55-W59.
- MI Davis, Jeremy P Hunt, Sanna Herrgard, Pietro Ciceri, Lisa M Wodicka, et al. (2011) Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol 29(11): 1046-1051.
- T Anastassiadis, SW Deacon, K Devarajan, H Ma, JR Peterson (2011) Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol 29(11): 1039-1045.
- L Wu, N Ai, Y Liu, Y Wang, X Fan (2013) Relating anatomical therapeutic indications by the ensemble similarity of drug sets. J Chem Inf Model 53(8): 2154-2160.
- L Chen, WM Zeng, YD Cai, KY Feng, KC Chou (2012) Predicting anatomical therapeutic chemical (ATC) classification of drugs by integrating chemical-chemical interactions and similarities. PLoS One 7(4): e35254.
- H Gurulingappa, C Kolárik, M Hofmann Apitius, J Fluck (2009) Concept-based semi-automatic classification of drugs. J Chem Inf Model 49(8): 1986-1992.
- X Cheng, SG Zhao, X Xiao, KC Chou (2017) iATC-mHyb: a hybrid multi-label classifier for predicting the classification of anatomical therapeutic chemicals. Oncotarget 8(35): 58494-58503.
- X Cheng, S G Zhao, X Xiao, and K C Chou, (2017) iATC-mISF: a multi-label classifier for predicting the classes of anatomical therapeutic chemicals. Bioinformatics 33(3): 341-346.
- Y Wang, Stephen H Bryant, Tiejun Cheng, Jiyao Wang, Asta Gindulyte, et al. (2016) PubChem BioAssay: 2017 update. Nucleic Acids Res 45(D1): D955-D963.
- D Mendez, Anna Gaulton, A Patrícia Bento, Jon Chambers, Marleen De Veij, et al. (2019) ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res 47(D1): D930-D940.
- JL Durant, BA Leland, DR Henry, JG Nourse (2002) Reoptimization of MDL keys for use in drug discovery. J Chem Inf Comput Sci 42(6): 1273-1280.
- D Rogers, M Hahn (2010) Extended-connectivity fingerprints. J Chem Inf Model 50(5): 742-754.
- DJ Mason, Ian Stott, Stephanie Ashenden, Zohar B Weinstein, Idil Karakoc, et al. (2017) Prediction of antibiotic interactions using descriptors derived from molecular structure. J Med Chem 60 (9): 3902-3912.
- G Landrum (2006) RDKit: Open-source cheminformatics.
- TK Ho (1995) Random decision forests. in Proceedings of 3rd international conference on document analysis and recognition 1: 278-282.
- L Breiman (2001) Random forests. Mach Learn 45(1): 5-32.
- G Biau (2012) Analysis of a random forests model. The Journal of Machine Learning Research 13(1): 1063-1095.
- L Patel, T Shukla, X Huang, D W Ussery, S Wang (2020) Machine learning methods in drug discovery. Molecules 25 (22): 5277.
- AP Lind, PC Anderson (2019) Predicting drug activity against cancer cells by random forest models based on minimal genomic information and chemical properties. PLoS One 14(7): e0219774.
- K Lee, M Lee, D Kim (2017) Utilizing random Forest QSAR models with optimized parameters for target identification and its application to target-fishing server. BMC Bioinformatics 18(16): 75-86.
- P Probst, M N Wright, A Boulesteix (2019) Hyperparameters and tuning strategies for random forest. Wiley Interdiscip Rev Data Min Knowl Discov 9(3): e1301.
- S Wang, X Yao (2012) Multiclass imbalance problems: Analysis and potential solutions. IEEE Transactions on Systems, Man, and Cybernetics, Part B (Cybernetics) 42(4): 1119-1130.
- NV Chawla, KW Bowyer, LO Hall, WP Kegelmeyer (2002) SMOTE: synthetic minority over-sampling technique. Journal of artificial intelligence research 16: 321-357.
- G Idakwo, Sundar Thangapandian, Joseph Luttrell, Yan Li, Nan Wang, et al. (2020) Structure-activity relationship-based chemical classification of highly imbalanced Tox21 datasets. J Cheminform 12(1): 1-19.
- LM Taft, RS Evans, CR Shyu, MJ Egger, N Chawla, et al. (2009) Countering imbalanced datasets to improve adverse drug event predictive models in labor and delivery. J Biomed Inform 42(2): 356-364.
- J Bergstra, Y Bengio (2012) Random search for hyper-parameter optimization. Journal of machine learning research 13(2).
- K Takahashi, K Yamamoto, A Kuchiba, T Koyama (2022) Confidence interval for micro-averaged F1 and macro-averaged F1 scores. Appl Intell 52(5): 4961-4972.




 We use cookies to ensure you get the best experience on our website.
We use cookies to ensure you get the best experience on our website.